Deployment Metadata
Overview
Teaching: 90 min
Exercises: 0 minQuestions
How do I load new deployments into the Database?
Objectives
Understand how to complete the template
Understand how to use the GitLab checklist
Learn how to use the
Deploynotebook
Process workflow
The process workflow for deployment metadata is as follows:
flowchart LR
tag_start(( )) --> get_meta(Receive
deployment metadata
from researchers)
style tag_start fill:#00FF00,stroke:#00FF00,stroke-width:4px
get_meta --> gitlab(Create
Gitlab
issue)
gitlab --> inspect(Visually
inspect)
inspect --> nodebook(Process and verify
with nodebooks)
nodebook --> plone(Add metadata
to repository folder)
plone --> otn(Pass to
OTN)
otn --> end2(( ))
style end2 fill:#FF0000,stroke:#FF0000
Once a project has been registered, the next step (for Deployment and Data project types) is to begin to quality control and load the instrument deployment metadata into the database. Deployment metadata should be reported to the Node in the template provided here. This file will contain information about the deployment of any instruments used to detect tagged subjects or collect related data. This includes stationary test tags, range test instruments, non-acoustic environmental sensors, and so on. Geographic location is recorded, as well as the duration of the deployment for each instrument. The locations of these listening stations are used to fix detections geographically.
Recall that there are multiple levels of data-tables in the database for deployment records: raw tables, rcvr_locations, stations and moorings. The process for loading instrument metadata reflects this, as does the GitLab task list.
Submitted Metadata
Immediately upon receipt of the metadata, create a new GitLab issue. Please use the Receiver_metadata Issue checklist template.
Here is the Issue checklist, for reference:
Receiver Metadata
- [ ] - NAME add label *'loading records'*
- [ ] - NAME load raw receiver metadata (`deploy` notebook) **put_table_name_in_ticket**
- [ ] - NAME check that station locations have not changed station "NAMES" since last submission (manual check)
- [ ] - NAME verify raw table (`deploy` notebook)
- [ ] - NAME post updated metadata file to project repository (OTN members.oceantrack.org, FACT RW etc)
- [ ] - NAME email notification of updated metadata file to PI and individual who submitted
- [ ] - NAME load station records (`deploy` notebook)
- [ ] - NAME verify stations (`deploy` notebook)
- [ ] - NAME load to rcvr_locations (`deploy` notebook)
- [ ] - NAME verify rcvr_locations (`deploy` notebook)
- [ ] - NAME add transmitter records receivers with integral pingers (`deploy` notebook)
- [ ] - NAME load to moorings (`deploy` notebook)
- [ ] - NAME verify moorings (`deploy` notebook)
- [ ] - NAME label issue with *'Verify'*
- [ ] - NAME pass issue to OTN DAQ for reassignment to analyst
- [ ] - NAME check if project is OTN loan, if yes, check for lost indicator in recovery column, list receiver serial numbers for OTN inventory updating.
- [ ] - NAME pass issue to OTN analyst for final verification
- [ ] - NAME check for double reporting (verification_notebooks/Deployment Verification notebook)
**receiver deployment files/path:**
Visual Inspection
Once the researcher provides the completed file, the Data Manager should complete a visual check for formatting and accuracy.
In general, the deployment metadata contains information on the instrument, the deployment location, and the deployment/recovery times.
Check for the following in the deployment metadata:
- Is there any information missing from the essential columns? These are:
- otn_array
- station_no
- deploy_date_time
- deploy_lat
- deploy_long
- ins_model_no
- ins_serial_no
- recovered
- recover_date_time
- If any of the above mandatory fields are blank, follow-up with the researcher will be required if:
- you cannot discern the values yourself.
- you do not have access to the Tag or Receiver Specifications from the manufacturer (relevant for the columns containing transmitter information).
- Are the station names in the metadata consistent with those already loaded to the database (ex. ‘_yyyy’ appended to station names or special characters in the metadata)?
- Are all lat/longs in the correct sign? Are they in the correct format (decimal degrees)?
- Do all transceivers/test tags have their transmitters provided?
- Are all recoveries from previous years recorded?
- Do comments suggest anything was lost or damaged, where recovery indicator doesn’t say “lost” or “failed”?
In general, the most common formatting errors occur in records where there are >1 instrument deployed at a station, or where the receiver was deployed and recovered from the same site.
The metadata template available here has a Data Dictionary sheet which contains detailed expectations for each column. Refer back to these definitions often. We have also included some recommendations on our FAQ page. Here are some guidelines:
- Deployment, download, and recovery information for each station is entered on a single line.
- When more than one instrument is deployed, downloaded, or recovered at the same station, enter each one on a separate line using the same
OTN_ARRAYandSTATION_NO. - When sentinel tags are co-deployed with receivers, their information can be added to
TRANSMITTERandTRANSMIT_MODELcolumns, on the same line as the receiver deployment. - If a sentinel tag is deployed alone then a new line for that station, with as much information as possible, is added.
- When stations are moved to a new location, but the researcher wants to keep the same station names, we often recommend appending ‘_yyyy’ to the station name, but this change might be forgotten the next time they submit metadata. So, we need to manually compare between the database and the metadata for special cases like this. Researchers may also submit station names with special characters which have been previously corrected and loaded to the database We need to make sure those same changes are reflected in the new metadata.
- When an instrument is deemed lost, a value of
lorlostshould be entered in the “recovered” field; if the instrument is found, this can be updated by changing the recovery field toforfoundand resubmitting the metadata sheet. - Every time an instrument is brought to the surface, enter
yto indicate it was successfully recovered, even if only for downloading and redeployment. A new line for the redeployment is required.
Task List Checkpoint
In GitLab, this task can be completed at this stage:
- [ ] - check that station locations have not changed station "NAMES" since last submission (manual check)
Quality Control - Deploy Nodebook
Each step in the Issue checklist will be discussed here, along with other important notes required to use the Nodebook.
Imports Cell
This section will be common for most Nodebooks: it is a cell at the top of the notebook where you will import any required packages and functions to use throughout the notebook. It must be run first, every time.
There are no values here which need to be edited.
Path to File
In this cell, you need to paste a filepath to the relevant Deployment Metadata file. The filepath will be added between the provided quotation marks.
Correct formatting looks something like this:
# Shortfrom metadata path (xls, csv)
filepath = r'C:/Users/path/to/deployment_metadata.xlsx'
You also must select the format of the Deployment metadata. Currently, only the FACT Network uses a slightly different format than the template available here. If its relevant for your Node, you can edit the excel_fmt section.
Correct formatting looks something like this:
excel_fmt = 'otn' # Deployment metadata format 'otn' or 'fact'
Once you have added your filepath and chosen your template format, you can run the cell.
Next, you must choose which sheet you would like to quality control. Generally, it will be named Deployment but is often customized by researchers. Once you have selected the sheet name, do not re-run the cell to save the output - simply ensure the correct sheet is highlighted and move onto the next cell.
Table Name and Database
You will have to edit three sections:
schema = 'collectioncode'- Please edit to include the relevant project code, in lowercase, between the quotes.
table_name = 'c_shortform_YYYY_mm'- Within the quotes, please add your custom table suffix. We recommend using
year_monthor similar, to indicate the most-recently deployed/recovered instrument in the metadata sheet.
- Within the quotes, please add your custom table suffix. We recommend using
engine = get_engine()- Within the open brackets you need to open quotations and paste the path to your database
.kdbxfile which contains your login credentials. - On MacOS computers, you can usually find and copy the path to your database
.kdbxfile by right-clicking on the file and holding down the “option” key. On Windows, we recommend using the installed software Path Copy Copy, so you can copy a unix-style path by right-clicking. - The path should look like
engine = get_engine('C:/Users/username/Desktop/Auth files/database_conn_string.kdbx').
- Within the open brackets you need to open quotations and paste the path to your database
Once you have added your information, you can run the cell. Successful login is indicated with the following output:
Auth password:········
Connection Notes: None
Database connection established
Connection Type:postgresql Host:db.for.your.org Database:your_db_name User:your_node_admin Node:Node
Verification of File Contents
Run this cell to complete the first round of Quality Control checks.
The output will have useful information:
- Is the sheet formatted correctly? Correct column names, datatypes in each column, etc.
- Compared to the
stationstable in the database, are the station names correct? Have stations “moved” location? Are the reported bottom_depths significantly different (check for possibleftvsmvsftmerrors). - Are all recovery dates after the deployment dates?
- Are all the provided
ins_model_novalues present in theobis.instrument_modelstable? If not, please check the records in theobis.instrument_modelsand the source file to confirm there are no typos. If this is a new model which has never been used before, use theadd instrument_modelsNodebook to add the new instrument model. -
Do all transceivers/test tags have their transmitters provided? Do these match any manufacturer specifications we have in the database? Note: For VR2AR and VR2Tx type receivers, researcher can record internal transmitters under the ‘transmitter’ column (example format: A69-1303-12345). We will then associate these ‘detections’ with the receiver! Though it’s not a compulsory information, it does help us distinguish real and tesing detections for your project, which can reduce the risk of mismatching.
- Are there any overlapping deployments (one serial number deployed at multiple locations for a period of time)?
- Are all the deployments within the Bounding Box of the project. If the bounding box needs to be expanded to include the stations, you can use the
Square Draw Toolto re-draw the bounding box until you are happy with it. Once all stations are drawn inside the bounding box, press theAdjust Bounding Boxbutton to save the results. - Are there possible gaps in the metadata, based on previously-loaded
detectionsfiles? This will be investigated in theDetections-3bNodebook if you need more details.
The Nodebook will indicate the sheet has passed quality control by adding a ✔️green checkmark beside each section. The Nodebook will also generate an interactive plot for you to explore, summarizing the instruments deployed over time, and a map of the deployments.
Using the map, please confirm the following:
- The instrument deployment locations are in the part of the world expected based on the project abstract. Ex: lat/long have correct +/- signs.
- The instrument deployments do not occur on land.
If there is information that fails quality control, you should fix the source-file (potentially after speaking to the researcher) and try again.
Loading the Raw Table
ONLY once the source file has successfully passed ALL quality control checks can you load the raw table to the database.
You have already named the table above, so there are no edits needed in this cell.
The Nodebook will indicate the success of the table-creation with the following message:
Reading file 'deployment_metadata.xlsx' as otn formatted Excel.
Table Loading Complete:
Loaded XXX records into table schema.c_shortform_YYYY_mm
Task List Checkpoint
In GitLab, this task can be completed at this stage:
- [ ] - load raw receiver metadata ("deploy" notebook) **put_table_name_in_ticket**
Ensure you paste the table name (ex: c_shortform_YYYY_mm) into the indicated section before you check the box.
Verify Raw Table
This cell will now complete the Quality Control checks of the raw table. This is to ensure the Nodebook loaded the records correctly from the Excel sheet.
The output will have useful information:
- Are there any duplicate records?
- Is there missing information in the
ar_model_noandar_serial_nocolumns? If so, ensure that it makes sense for receivers to be deployed without Acoustic Releases in this location (ex: diver deployed). - Do all transceivers/test tags have their transmitters provided? Do these match any manufacturer specifications we have in the database?
- Are there any non-numeric serial numbers? Do these make sense (ex: environmental sensors)?
- Are the deployments within the bounding box?
- Is the
recoveredcolumn completed correctly, based on thecommentsand therecovery_datecolumns? - Are there blank strings that need to be set to NULL? If so, press the
Set to NULLbutton in that cell.
The Nodebook will indicate the sheet had passed quality control by adding a ✔️green checkmark beside each section.
If there are any errors, go into database and fix the raw table directly, or contact the researcher and then fix the raw table.
Task List Checkpoint
In GitLab, this task can be completed at this stage:
- [ ] - NAME verify raw table ("deploy" notebook)
Find Raw Data Table in DB (schema.c_shortform_YYYY_MM)
- This table will includes all the OTN compulsory columns for receiver metadata as well as the ones the researcher includes. But only OTN compulsory columns are QCed.
- An example query: `select * from schema.c_shortform_2020_04 cs where ins_model_no ilike '%CTD%'`
Loading Stations Records
STOP - confirm there is no Push currently ongoing. If a Push is ongoing, you must wait for it to be completed before processing beyond this point
Only once the raw table has successfully passed ALL quality control checks can you load the stations information to the database stations table.
Running this cell will first check for any new stations to add, then confirm the records in the stations table matches the records in the moorings table where basisofrecord = 'STATION'.
If new stations are identified:
- Compare these station names to existing stations in the
stationstable in the database. Are they truly new, or is there a typo in the raw table? Did the station change names, but it is in the same location as a previous station? - If there are fixes to be made, change the records in the raw table, or contact the researcher to confirm.
- If all stations are truly new, you should review the information in the editable form, then select
Add New Stations.
The success message will look like:
Adding station records to the stations table.
Creating new stations...
Added XX new stations to schema.moorings
If the stations and moorings tables are not in sync, you will need to compare the two tables for differences and possibly update one or the other.
Task List Checkpoint
In GitLab, this task can be completed at this stage:
- [ ] - load station records ("deploy" notebook)
Verify Stations Table
This cell will now complete the Quality Control checks of the stations records contained in the entire schema. We are no longer only checking against our newly-loaded records, but also each previously-loaded record in this schema/project. This will help catch historical errors.
The output will have useful information:
- Were all the stations from our raw table promoted to the
stationstable and themooringstable? - Are all stations in unique locations?
- Are all stations within the project’s bounding box?
- Does the date in the
stationstable match the first deployment date for that station? - Are there blank strings that need to be set to NULL? If so, press the
Set to NULLbutton in that cell. - Are any of the dates in the future?
The Nodebook will indicate the sheet had passed quality control by adding a ✔️green checkmark beside each section.
If there are any errors, you could directly connect to the database and fix the raw table directly, or contact the researcher and then fix the raw table using updated input metadata. If there are problems with records that have already been promoted to the stations or moorings tables, you will need to create a db fix ticket in Gitlab in order to correct the records in the database.
Task List Checkpoint
In GitLab, this task can be completed at this stage:
- [ ] - verify stations ("deploy" notebook)
Find Station Tables in DB(schema.stations & schema.rcvr_locations)
- These Station tables are intermediate tables. They grab the necessary information from the raw table.
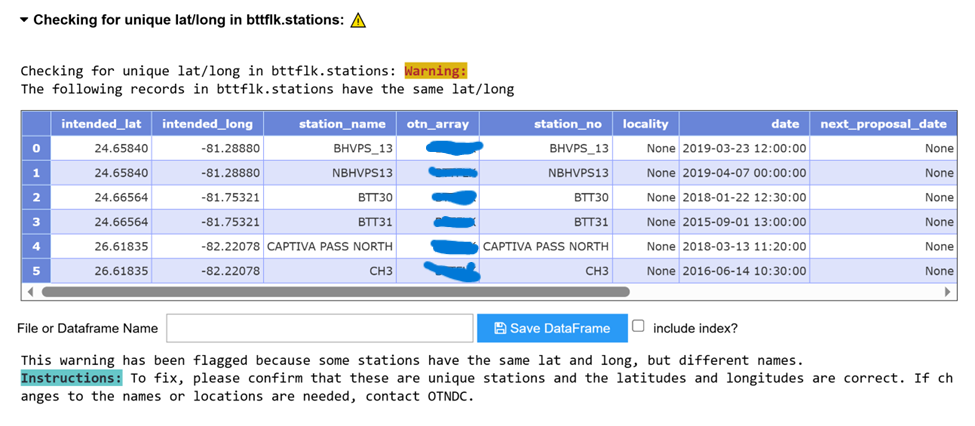
- `schema.stations` contains all distinct the deployment stations information from that schema across all years. Column `date` and `intended_lon`, `intended_lat` represent the first time and coordinate (lon,lat) this station was added.
- Note: In `schema.stations` table, all stations have the distinct names with distinct coordinates. The notebook will show errors, if the researcher put different coordinates for the same location or put the same coordinate for different locations. Here we can use corresponding DB fix tool to change station names.
- An example query: `select * from schema.stations where station_name in ('A','B')`
Load to rcvr_locations
Once the station table is verified, the receiver deployment records can now be promoted to the “intermediate” rcvr_locations table.
The cell will identify any new deployments to add and any previously-loaded deployments which need updating (ex: they have been recovered).
If new deployments are identified:
- Compare these deployments to existing deployments in the
rcvr_locationstable in the database. Are they truly new, or is there a typo in the raw table? Did the station change names, but it is in the same location as before? Did the serial number change, but the deployment location and date are the same? - If there are fixes to be made, change the records in the
rawtable or contact the researcher. - If all deployments are truly new, you can then select
Add Deployments.
If deployment updates are identified:
- Review the suggested changes: do not select changes which remove information (ex: release 590023 –>
Noneis not a good change to accept). - Use the check boxes to select only the appropriate changes.
- Once you are sure the changes are correct, you can then select
Update Deployments.
Each instance will give a success message such as:
Loading deployments into the rcvr_locations_table
Loaded XX records into the schema.rcvr_locations table.
Task List Checkpoint
In GitLab, this task can be completed at this stage:
- [ ] - load to rcvr_locations ("deploy" notebook)
Verify rcvr_locations
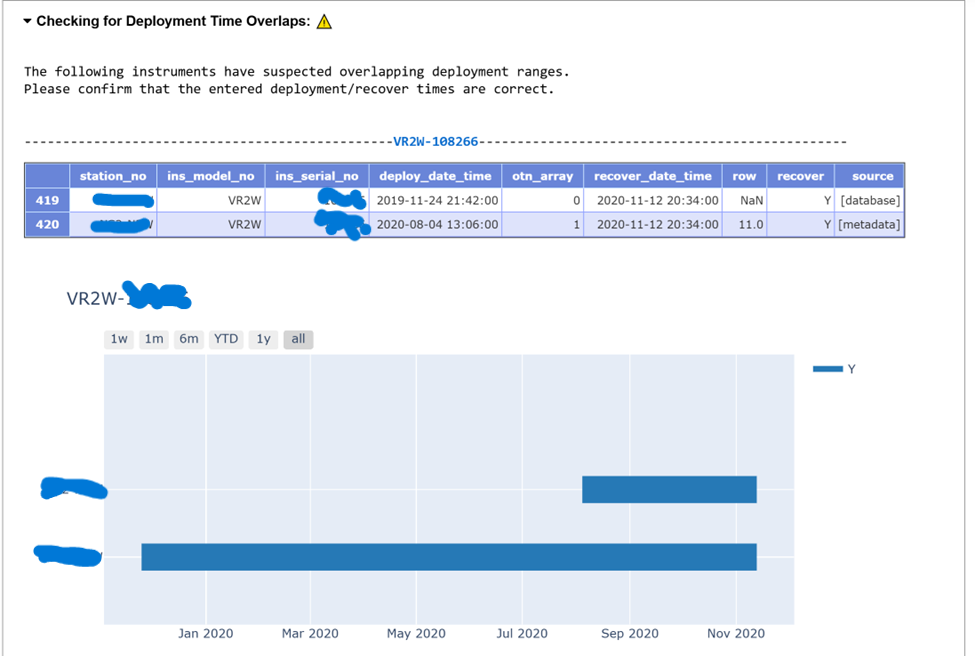
This cell will now complete the Quality Control checks of the rcvr_locations records contained in the entire schema. We are no longer only checking our newly-loaded records, but also each previously-loaded record for this schema/project. This will help catch historical errors.
The output will have useful information:
- Have all deployments been loaded from the raw table? Please note that instruments where a sentinel tag is deployed alone at a station will not be loaded to rcvr_locations, and so these will likely be flagged in this section for your review.
- Are there blank strings that need to be set to NULL? If so, press the
Set to NULLbutton in that cell. - Based on comments, are there any instances where a receiver’s status should be changed to “lost”?
- Are all instruments in the
obis.instrument_modelstable? - Are there any overlapping deployments?
- Are
the_geom,serialnumber, andcatalognumberformatted correctly?
The Nodebook will indicate the table has passed quality control by adding a ✔️green checkmark beside each section.
If there are any errors with records that have already been promoted to the rcvr_locations table, you will need to create a db fix ticket in Gitlab to correct the records in the database. You may need to contact the researcher before resolving the error.
Task List Checkpoint
In GitLab, this task can be completed at this stage:
- [ ] - verify rcvr_locations ("deploy" notebook)
Find schema.rcvr_locations tables in DB
- These tables are also imtermediate tables which contains all deployment information for all receivers from that schema across all years. Station is treated as an area, receivers can be deployed at the same station with different coordinates. Column
deploy_dateanddeploy_lon,deploy_latshows each receiver’s deployment date and coordinates.- Note: In
schema.rcvr_locationstable, you may see the same station has different coordinates. But the notebook will show errors if the same receiver’s overlapping deployments. Here we can look and this table for information we need to change and use corresponding DB fix tool. - An example query:
select * from schema.rcvr_locations rl where rl.rcv_serial_no = 'XXXXX'
- Note: In
Load Transmitter Records to Moorings
The transmitter values associated with transceivers, co-deployed sentinel tags, or stand-alone test tags will be loaded to the moorings table in this section. Existing transmitter records will also be updated, if relevant.
If new transmitters are identified:
- Review for accuracy then select
Add Transmitters.
If transmitter updates are identified:
- Review the suggested changes: do not select changes which remove information (ex: release 590023 –>
Noneis not a good change to accept). - Use the check boxes to select only the appropriate changes.
- Once you are sure the changes are correct, you can then select
Update Transmitters.
Task List Checkpoint
In GitLab, this task can be completed at this stage:
- [ ] - load transmitter records receivers with integral pingers ("deploy" notebook)
Load Receivers to Moorings
The final, highest-level table for instrument deployments is moorings.
The cell will identify any new deployments to add and any previously-loaded deployments which need updating (ex: they have been recovered).
Please review all new deployments and deployment updates for accuracy, then press the associated buttons to make the changes. At this stage, the updates are not editable: any updates chosen from the rcvr_locations section will be processed here.
You may be asked to select an instrumenttype for certain receivers. Use the drop-down menu to select before adding the deployment.
Task List Checkpoint
In GitLab, this task can be completed at this stage:
- [ ] - load to moorings ("deploy" notebook)
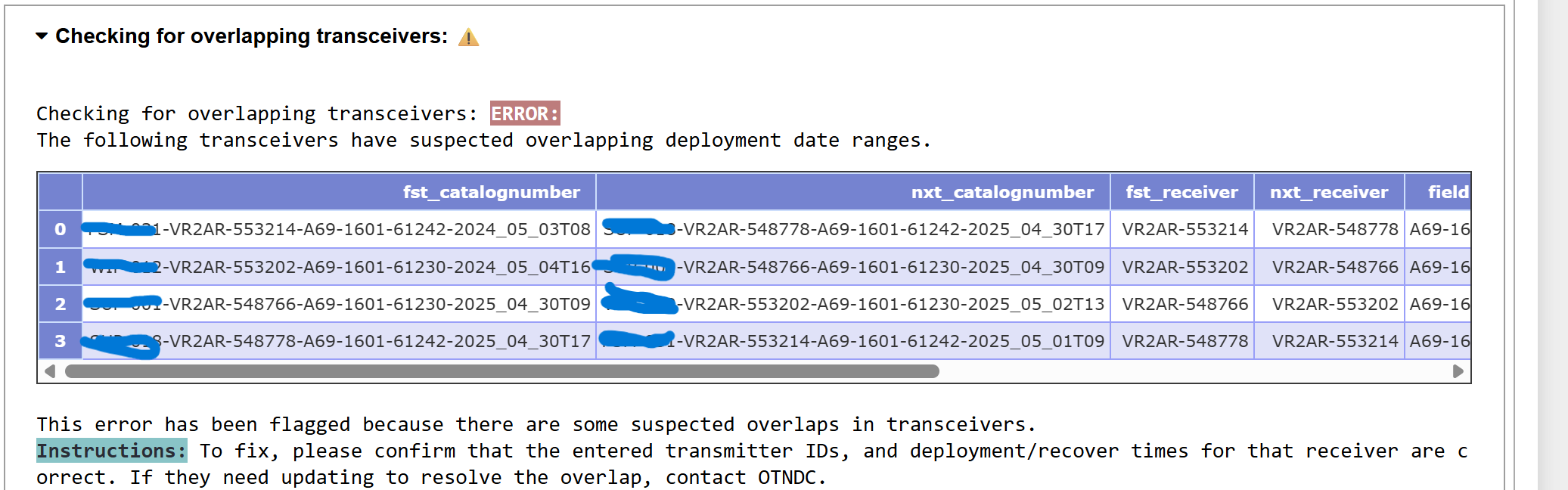
Verify Moorings
This cell will now complete the Quality Control checks of the moorings records contained in the entire schema. We are no longer only checking our newly-loaded records, but also each previously-loaded record in this project/schema.
The output will have useful information:
- Have all deployments been loaded from rcvr_locations?
- Do we have manufacturer specifications for the receivers or tags deployed?
- Are there blank strings that need to be set to NULL? If so, press the
Set to NULLbutton in that cell. - Do the
STATIONSrecords match the information in thestationstable? - Are all instruments in the
obis.instrument_modelstable? - Are there any overlapping deployments or receivers or tags?
- Are there duplicate download records?
- Are the
latitude,longitude,the_geom,serialnumber, andcatalognumberformatted correctly?
The Nodebook will indicate the table has passed quality control by adding a ✔️ green checkmark beside each section.
If there are any errors with records that have already been promoted to the moorings table, you will need to create a db fix ticket in Gitlab to correct the records in the database. You may need to contact the researcher before resolving the error.
Find Mooring Tables in DB
- `schema.moorings` contains all receiver, transmitter, event information from this project. Note: the notebook will show errors if the same transmitter_ID has been used in different receivers. We can check this table to check more information on transmitter_ID and may need to use the corresponding DB fix tool to change transmitter_ID.
- An example query: `select * from schema.moorings where basisofrecord = 'TRANSMITTER' and relationshiptype = 'STATION'`
Task List Checkpoint
In GitLab, this task can be completed at this stage:
- [ ] - verify moorings ("deploy" notebook)
Final Steps
The remaining steps in the GitLab Checklist are completed outside the Nodebooks.
First: you should access the Repository folder in your browser and add the cleaned Deployment Metadata .xlsx file into the “Data and Metadata” folder.
Finally, the GitLab ticket can be reassigned to an OTN analyst for final verification in the database.
Key Points
Loading receiver metadata requires judgement from the Data Manager
Communication with the researcher is essential when errors are found