Basic Visualization and Plotting with glatos
Overview
Teaching: 30 min
Exercises: 0 minQuestions
How can I use glatos to plot my data?
What kinds of plots can I make with my data?
Objectives
Note to instructors: please choose the relevant Network below when teaching
ACT Node
Now that we’ve cleaned and processed our data, we can use glatos’ built-in plotting tools to make quick and effective visualizations out of it. One of the simplest visualizations is an abacus plot to display animal detections against the appropriate stations. To this end, glatos supplies a built-in, customizable abacus_plot function.
# Visualizing Data - Abacus Plots ####
# ?glatos::abacus_plot
# customizable version of the standard VUE-derived abacus plots
abacus_plot(detections_w_events,
location_col='station',
main='ACT Detections by Station') # can use plot() variables here, they get passed thru to plot()
This is good, but you can see that the plot is cluttered. Rather than plotting our entire dataset, let’s try filtering out a single animal ID and only plotting that. We can do this right in our call to abacus_plot with the filtering syntax we’ve previously covered.
# pick a single fish to plot
abacus_plot(detections_filtered[detections_filtered$animal_id== "CBCNR-1218508-2015-10-13",],
location_col='station',
main="CBCNR-1218508-2015-10-13 Detections By Station")
Other plots are available in glatos and can show different facets of our data. If we want to see the physical distribution of our stations, for example, a bubble plot will serve us better.
# Bubble Plots for Spatial Distribution of Fish ####
# bubble variable gets the summary data that was created to make the plot
detections_filtered
?detection_bubble_plot
# We'll use raster to get a polygon to plot against
library(geodata)
USA <- geodata::gadm("USA", level=1, path=".")
MD <- USA[USA$NAME_1=="Maryland",]
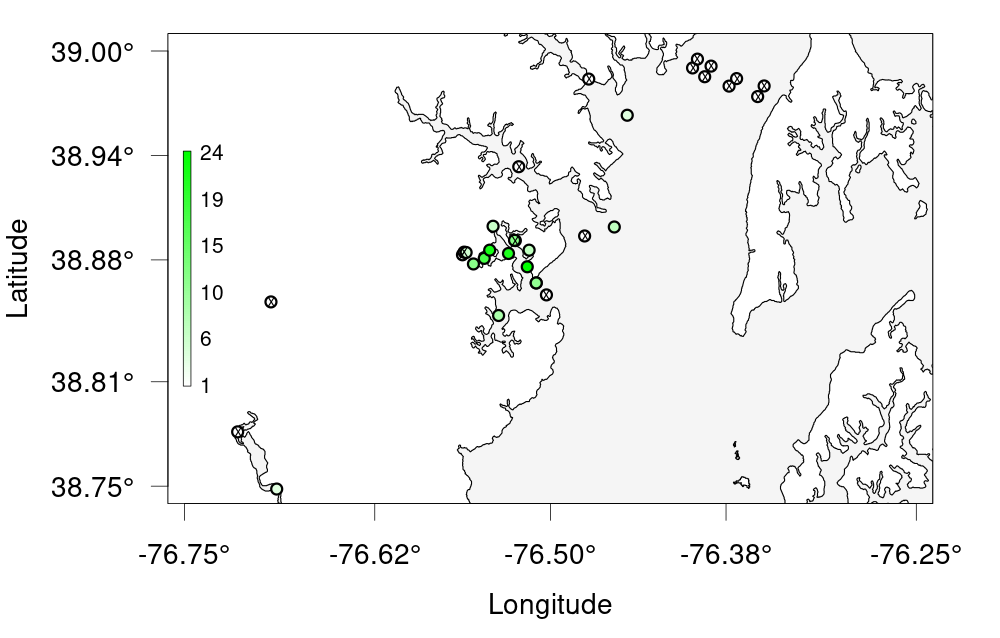
bubble_station <- detection_bubble_plot(detections_filtered,
background_ylim = c(38, 40),
background_xlim = c(-77, -76),
map = MD,
location_col = 'station',
out_file = 'act_bubbles_by_stations.png')
bubble_station
bubble_array <- detection_bubble_plot(detections_filtered,
background_ylim = c(38, 40),
background_xlim = c(-77, -76),
map = MD,
out_file = 'act_bubbles_by_array.png')
bubble_array
These examples provide just a brief introduction to some of the plotting available in glatos.
Glatos ACT Challenge
Challenge 1 —- Create a bubble plot of that bay we zoomed in earlier. Set the bounding box using the provided nw + se cordinates, change the colour scale and resize the points to be smaller. As a bonus, add points for the other receivers that don’t have any detections. Hint: ?detection_bubble_plot will help a lot Here’s some code to get you started
nw <- c(38.75, -76.75) # given se <- c(39, -76.25) # givenSolution
nw <- c(38.75, -76.75) # given se <- c(39, -76.25) # given deploys <- read_otn_deployments('matos_FineToShare_stations_receivers_202104091205.csv') # For bonus bubble_challenge <- detection_bubble_plot(detections_filtered, background_ylim = c(nw[1], se[1]), background_xlim = c(nw[2], se[2]), map = MD, symbol_radius = 0.75, location_col = 'station', col_grad = c('white', 'green'), receiver_locs = deploys, # For bonus out_file = 'act_bubbles_challenge.png')
FACT Node
Now that we’ve cleaned and processed our data, we can use glatos’ built-in plotting tools to make quick and effective visualizations out of it. One of the simplest visualizations is an abacus plot to display animal detections against the appropriate stations. To this end, glatos supplies a built-in, customizable abacus_plot function.
# Visualizing Data - Abacus Plots ####
# ?glatos::abacus_plot
# customizable version of the standard VUE-derived abacus plots
abacus_plot(detections_w_events,
location_col='station',
main='TQCS Detections by Station') # can use plot() variables here, they get passed thru to plot()
This is good, but you can see that the plot is cluttered. Rather than plotting our entire dataset, let’s try filtering out a single animal ID and only plotting that. We can do this right in our call to abacus_plot with the filtering syntax we’ve previously covered.
# pick a single fish to plot
abacus_plot(detections_filtered[detections_filtered$animal_id=="TQCS-1049273-2008-02-28",],
location_col='station',
main="TQCS-1049273-2008-02-28 Detections By Station")
Other plots are available in glatos and can show different facets of our data. If we want to see the physical distribution of our stations, for example, a bubble plot will serve us better.
# Bubble Plots for Spatial Distribution of Fish ####
# bubble variable gets the summary data that was created to make the plot
detections_filtered
?detection_bubble_plot
# We'll use raster to get a polygon to plot on
library(geodata)
USA <- geodata::gadm("USA", level=1, path=".")
FL <- USA[USA$NAME_1=="Florida",]
#Alternative method of getting the polygon.
f <- 'http://biogeo.ucdavis.edu/data/gadm3.6/Rsp/gadm36_USA_1_sp.rds'
b <- basename(f)
download.file(f, b, mode="wb", method="curl")
USA <- readRDS('gadm36_USA_1_sp.rds')
FL <- USA[USA$NAME_1=="Florida",]
bubble_station <- detection_bubble_plot(detections_filtered,
out_file = 'tqcs_bubble.png',
location_col = 'station',
map = FL,
col_grad=c('white', 'green'),
background_xlim = c(-81, -80),
background_ylim = c(26, 28))
bubble_station
These examples provide just a brief introduction to some of the plotting available in glatos.
Glatos FACT Challenge
Challenge 1 —- Create a bubble plot of that bay we zoomed in earlier. Set the bounding box using the provided nw + se cordinates, change the colour scale and resize the points to be smaller. As a bonus, add points for the other receivers that don’t have any detections. Hint: ?detection_bubble_plot will help a lot Here’s some code to get you started
nw <- c(26, -80) # given se <- c(28, -81) # givenSolution
nw <- c(26, -80) # given se <- c(28, -81) # given bubble_challenge <- detection_bubble_plot(detections_filtered, background_ylim = c(nw[1], se[1]), background_xlim = c(nw[2], se[2]), map = FL, symbol_radius = 0.75, location_col = 'station', col_grad = c('white', 'green'), receiver_locs = receivers, # For bonus out_file = 'fact_bubbles_challenge.png')
GLATOS Network
Now that we’ve cleaned and processed our data, we can use glatos’ built-in plotting tools to make quick and effective visualizations out of it. One of the simplest visualizations is an abacus plot to display animal detections against the appropriate stations. To this end, glatos supplies a built-in, customizable abacus_plot function.
# Visualizing Data - Abacus Plots ####
# ?glatos::abacus_plot
# customizable version of the standard VUE-derived abacus plots
abacus_plot(detections_w_events,
location_col='station',
main='Walleye detections by station') # can use plot() variables here, they get passed thru to plot()
This is good, but you can see that the plot is cluttered. Rather than plotting our entire dataset, let’s try filtering out a single animal ID and only plotting that. We can do this right in our call to abacus_plot with the filtering syntax we’ve previously covered.
# pick a single fish to plot
abacus_plot(detections_filtered[detections_filtered$animal_id=="22",],
location_col='station',
main="Animal 22 Detections By Station")
Other plots are available in glatos and can show different facets of our data. If we want to see the physical distribution of our stations, for example, a bubble plot will serve us better.
# Bubble Plots for Spatial Distribution of Fish ####
# bubble variable gets the summary data that was created to make the plot
?detection_bubble_plot
bubble_station <- detection_bubble_plot(detections_filtered,
location_col = 'station',
out_file = 'walleye_bubbles_by_stations.png')
bubble_station
bubble_array <- detection_bubble_plot(detections_filtered,
out_file = 'walleye_bubbles_by_array.png')
bubble_array
These examples provide just a brief introduction to some of the plotting available in glatos.
Glatos Challenge
Challenge 1 —- Create a bubble plot of the station in Lake Erie only. Set the bounding box using the provided nw + se cordinates and resize the points. As a bonus, add points for the other receivers in Lake Erie. Hint: ?detection_bubble_plot will help a lot Here’s some code to get you started
erie_arrays <-c("DRF", "DRL", "DRU", "MAU", "RAR", "SCL", "SCM", "TSR") #given nw <- c(43, -83.75) #given se <- c(41.25, -82) #givenSolution
erie_arrays <-c("DRF", "DRL", "DRU", "MAU", "RAR", "SCL", "SCM", "TSR") #given nw <- c(43, -83.75) #given se <- c(41.25, -82) #given bubble_challenge <- detection_bubble_plot(detections_filtered, background_ylim = c(nw[1], se[1]), background_xlim = c(nw[2], se[2]), symbol_radius = 0.75, location_col = 'station', col_grad = c('white', 'green'), out_file = 'glatos_bubbles_challenge.png')
MIGRAMAR Node
Now that we’ve cleaned and processed our data, we can use glatos’ built-in plotting tools to make quick and effective visualizations out of it. One of the simplest visualizations is an abacus plot to display animal detections against the appropriate stations. To this end, glatos supplies a built-in, customizable abacus_plot function.
# Visualizing Data - Abacus Plots ####
# ?glatos::abacus_plot
# customizable version of the standard VUE-derived abacus plots
abacus_plot(detections_w_events,
location_col='station',
main='MIGRAMAR Detections by Station') # can use plot() variables here, they get passed thru to plot()
This is good, but you can see that the plot is cluttered. Rather than plotting our entire dataset, let’s try filtering out a single animal ID and only plotting that. We can do this right in our call to abacus_plot with the filtering syntax we’ve previously covered.
# pick a single fish to plot
abacus_plot(detections_filtered[detections_filtered$animal_id== "GMR-25724-2014-01-22",],
location_col='station',
main="GMR-25724-2014-01-22 Detections By Station"))
Other plots are available in glatos and can show different facets of our data. If we want to see the physical distribution of our stations, for example, a bubble plot will serve us better.
# Bubble Plots for Spatial Distribution of Fish ####
# bubble variable gets the summary data that was created to make the plot
detections_filtered
?detection_bubble_plot
# We'll use raster to get a polygon to plot against
library(raster)
library(geodata)
ECU <- geodata::gadm("Ecuador", level=1, path=".")
GAL <- ECU[ECU$NAME_1=="Galápagos",]
bubble_station <- detection_bubble_plot(detections_filtered,
background_ylim = c(-2, 2),
background_xlim = c(-93.5, -89),
map = GAL,
location_col = 'station',
out_file = 'migramar_bubbles_by_stations.png')
bubble_station
These examples provide just a brief introduction to some of the plotting available in glatos.
Glatos MIGRAMAR Challenge
Challenge 1 —- Create a bubble plot of the islands we zoomed in earlier. Set the bounding box using the provided nw + se cordinates, change the colour scale and resize the points to be smaller. Hint: ?detection_bubble_plot will help a lot Here’s some code to get you started
nw <- c(-2, -89) # given se <- c(2, -93.5) # givenSolution
nw <- c(-2, -89) # given se <- c(2, -93.5) # given bubble_challenge <- detection_bubble_plot(detections_filtered, background_ylim = c(nw[1], se[1]), background_xlim = c(nw[2], se[2]), map = GAL, symbol_radius = 0.75, location_col = 'station', col_grad = c('white', 'green'), out_file = 'migramar_bubbles_challenge.png')
Key Points